GenBank Upload Pipeline 2: Geneious Prime GenBank Submission Tool
Installing the Geneious Plugin
Note
The most up-to-date GenBank Submission plugin version is 1.6.8, which can be found here.
The Geneious Prime’s GenBank Submission plugin tool utilizes BankIT to submit sequences to GenBank for all DNA barcode markers excluding metazoan (multicellular) COI or rRNA sequences such as 18S, 16S, 23S, 28S, ITS1, ITS2, etc. which must be submitted using the GenBank Upload Pipeline 1: GenBank Submission Portal.
The easiest way to install the plugin is from the Geneious Prime Menu Bar. Go to Tools > Plugins. A Preferences window will pop up. In the top “Available Plugins” section, scroll to the “GenBank Submission” plugin.
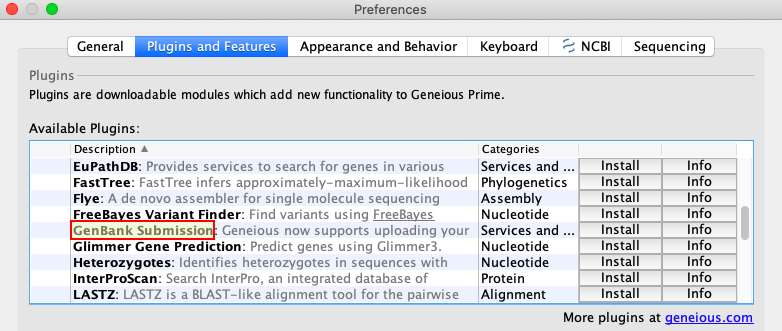
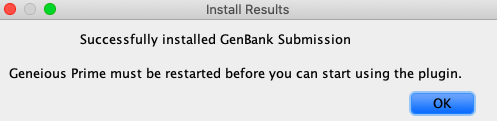
Click “Install”, and Geneious Prime will start downloading it from the Internet. If all goes well, a window will pop up to say the installation was successful, and that a restart will be needed. Click “OK” to restart Geneious Prime.
Using the Plugin
Please note, it is possible to submit to GenBank from either assembled contigs or consensus sequences. The directions laid out below will only describe submitting consensus sequences, as that is SIBN best practice.
Before starting a GenBank submission, organize all high quality consensus sequences to be submitted. Make sure the most up-to-date GEOME FIMS data and Biocode LIMS data are associated with the sequences. To do this, follow the instructions on the Annotating with FIMS/LIMS Data page to annotate the sequence files with the current GEOME FIMS and Biocode LIMS data.
Keep consensus sequences from different markers in separate folders as only one marker can be submitted at a time. It is possible to submit a single sample or a batch of samples at once. Also make sure to separate submissions based on organism type so that the correct genetic code can be selected for each submission.
Once ready, in the Geneious Prime Menu Bar, go to Tools > Submit to GenBank. A window will appear that has the following sections. Each section is detailed below.
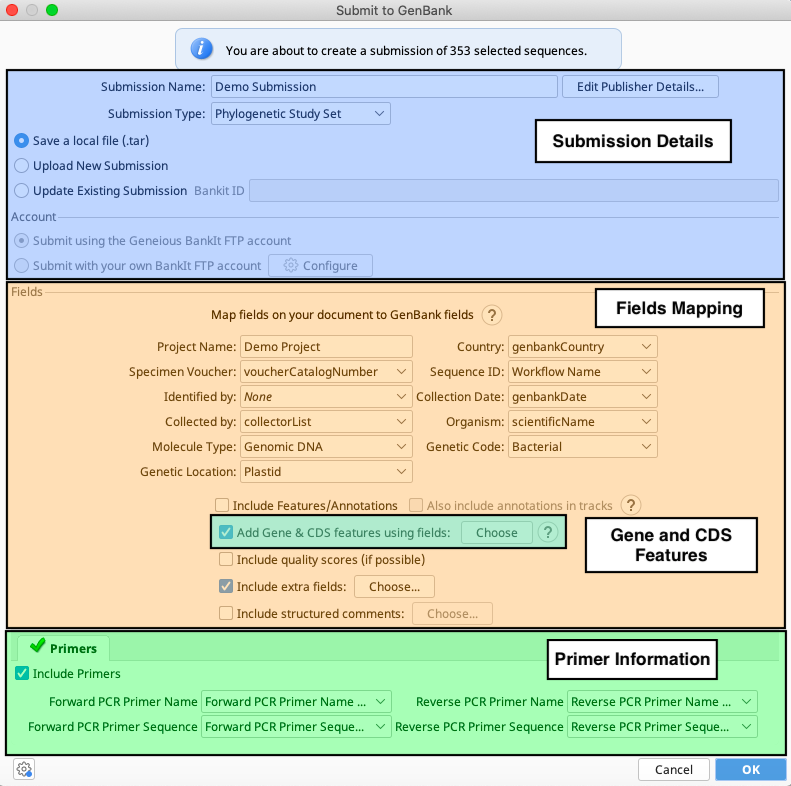
Submission Details
The first part of the GenBank submission deals with filling out the contact details and attributions for a sequence submission, as well as choosing submission type and how to submit to GenBank.
Give the “Submission Name” field a descriptive name. This entry will not show up in the GenBank record.
Choose the Submission Type based on the descriptions in the dropdown menu. This choice is enabled when submitting multiple sequences. In most projects for the SI Barcode Network, the submission type will be “Phylogenetic Study Set”.
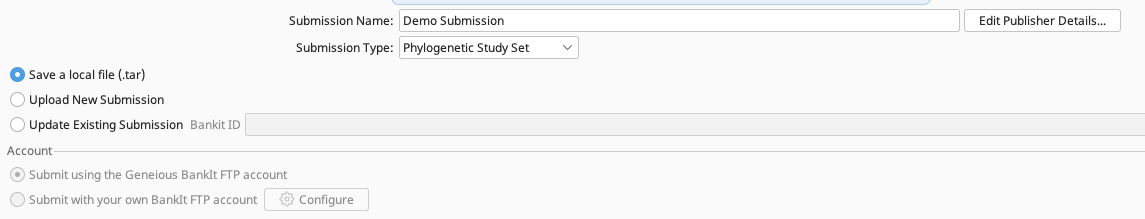
Then choose between “Save a local file (.tar)” or “Upload New Submission” to indicate how the submission will be packaged and sent to GenBank. User should choose which option, SIBN does not recommend one over the other.
Differences between the two submission options:
Save a local file (.tar) - A zipped file is produced through tbl2asn that must be emailed by the user to the GenBank submission team at gb-sub@ncbi.nlm.nih.gov.
Upload New Submission - Sequences will be bundled and sent directly to GenBank through the Geneious Prime BankIt ID.
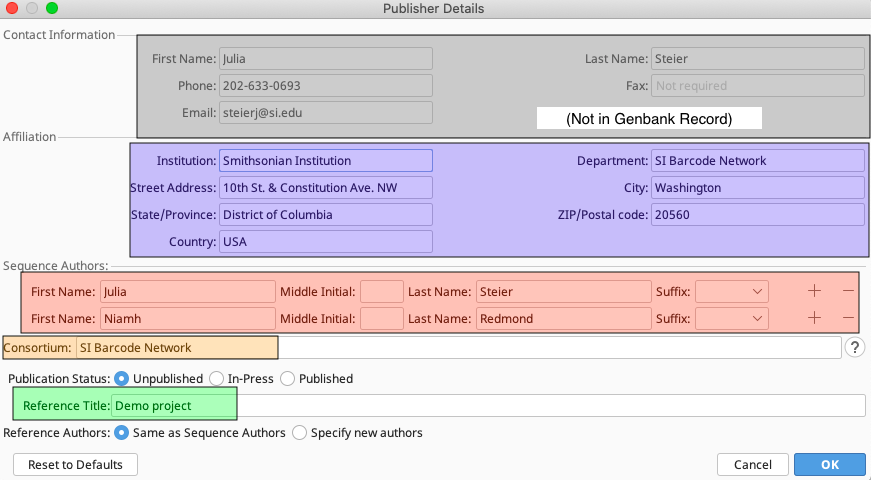
Click on the “Edit Publisher Details…” button to bring up the Publisher Details dialog box.
- Contact Information
Fill out the top section with the contact information for the submission. This will be the information that GenBank staff will use to contact the submitter with questions or updates for the submission.
- Affiliation
Fill out the relevant information for the institution that produced these sequences. The entries in this section will show up in the GenBank record, so be sure to provide accurate and consistent information.
- Sequence Authors
List any individuals who were involved in the production of these sequences, and particularly anyone who may need to make edits in the future to the GenBank records. Keep in mind that only those listed on the record will be authorized to make changes to the GenBank record. If a publication is or will be associated with these sequences, make sure to include at least one publication author as a sequence author.
- Consortium
This is an optional field that links the records to any group that may be associated with the sequence data. However, only individuals who are listed as Sequence Authors have authority to edit GenBank records.
- Publication Status and Title
Since the SI Barcode Network attempts to get sequence records published to GenBank as quickly as possible, there will generally not be an associated publication yet. For cases like these, select “Unpublished” for the Publication Status, and the project name for the Publication Title. This allows for easier searching and filtering, and publications can be added to sequences later as they are published.
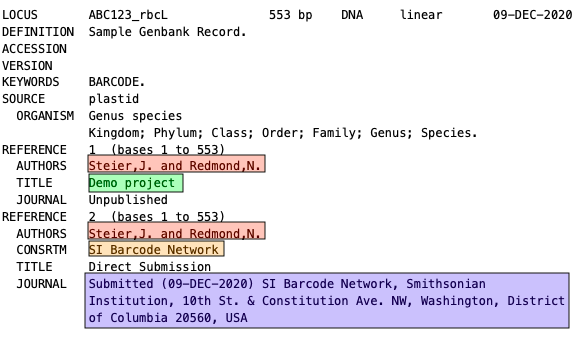
This portion of a sample GenBank record shows how the different parts of the Publisher Details section will appear when the record is published.
Fields Mapping
The next part of the Submit to GenBank window will be used to map all of the different specimen metadata fields to the GenBank record.
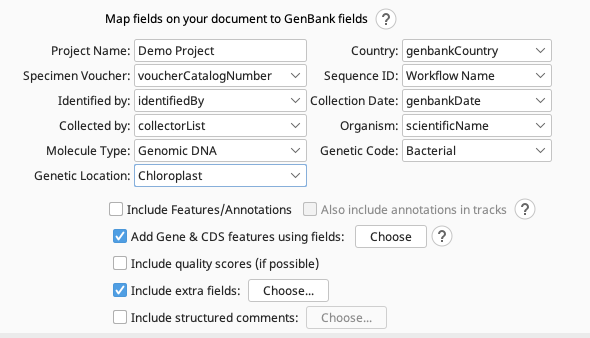
This screenshot shows the appropriate FIMS fields to select for each of the GenBank fields.
- Project Name
Just like the “Submission Name” field at the beginning, this entry won’t end up in the GenBank record, but should be a meaningful name used to organize the sequences.
- Country
This will become the “country” field in GenBank. This corresponds to the field “genbankCountry’’ found in the Geneious LIMS data. Within the GEOME FIMS, this is separated into “Country” and “Locality” fields so that the “Country” value can be validated according to the INSDC country list (http://www.insdc.org/country.html). Geneious should automatically combine these two fields into the field “genbankCountry”, if the sequences are annotated correctly.
- Specimen Voucher
This will become the “specimen_voucher” field in GenBank. It corresponds with the GEOME FIMS field “voucherCatalogNumber”, which should be a colon-separated triplet consisting of [institutionCode]:[collectionCode]:[catalogNumber]. If the voucher is from a botanical collection, the voucherCatalogNumber should be a doublet consisting of [herbariumCode]:[catalogNumber] or [collector surname]:[collector number].
- Sequence ID
This field will not be published as part of the GenBank record, but it is very important because this field will connect the specimen data and sequence data. Select the LIMS field “Workflow Name” for this.
- Identified by
This will become the “identified_by” field in GenBank. It corresponds with the GEOME FIMS field “identifiedBy”. If it is unknown, select None.
- Collection Date
This will become the “collection_date” field in GenBank. In GEOME FIMS, this is separated into “yearCollected”, “monthCollected”, and “dayCollected” fields so that each could be validated. However, Geneious should automatically combine these fields into one “genbankDate” field if the sequences are annotated correctly.
- Collected by
This will become the “collected_by” field in GenBank. It corresponds with the GEOME FIMS field “collectorList”. If it is unknown, select None.
- Organism
This field corresponds with the “scientificName” field from GEOME FIMS. It will be checked against the NCBI taxonomy database, so if it is not already in the database, NCBI staff will create a new entry in the database. This will be the case with any names not identified to species and any morphospecies. The name should only be the binomial name (or trinomial if subspecies), and should not include the taxonomic name authority.
- Molecule Type
This will always be “Genomic DNA” for DNA Barcode records.
- Genetic Code
For animal cytB barcode sequences, this will be either “Vertebrate Mitochondrial” or “Invertebrate Mitochondrial”. Plant barcode sequences (matK and rbcL) will always be “Bacterial” (the full name that Geneious abbreviates is “The Bacterial, Archaeal, and Plant Plastid Code”). For non-coding sequences, Genetic Code may be left on the default “Standard” and will not be used.
- Genetic Location
For cytB sequences, this will be “Mitochondrion”. For plant barcode sequences (matK, rbcL, and psbA-trnH - a common secondary barcode region), this will be “Chloroplast”. For any nuclear derived regions, this will be “Genomic”.
Include Extra Fields
If there is any extra collection information that should be included in these GenBank records, it can be added by checking the “Include extra fields” option below the set Fields discussed above. Click “Choose” and the Choose Additional Fields window should appear with dropdown menu options.
The most common GenBank field to add from the Field Name menu is “Lat_Lon”. If latitude and longitude data are available in the GEOME FIMS in separate “decimalLatitude” and “decimalLongitude’’ fields, Geneious should automatically combine these fields into “genbankLatLng” which can be found in the “Field Value” dropdown menu.
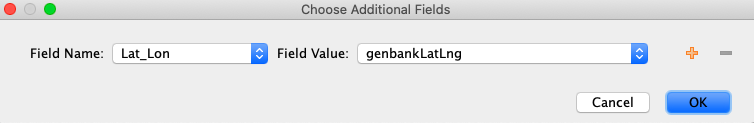
Examples of other fields that might be used here from the Field Name dropdown menu are “Host”, “Isolate”, or “Bio_material”, depending on the nature of the samples and metadata. The “Notes” GenBank field can be added if notes on the taxonomic confidence kept in the LIMS field “Assembly Notes” need to be added to the GenBank records.
Gene and CDS Features
When submitting protein-coding sequences, the next step will be to indicate which protein-coding gene was sequenced. As seen in the snippet from a sample GenBank record below, this will also provide enough information for Geneious to automatically generate the protein amino acid sequence as well.
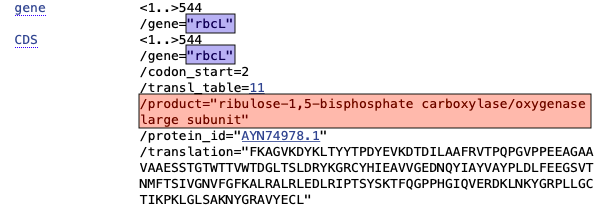
Since DNA barcodes are not full gene sequences, select “Partial” for both Gene Feature and CDS Feature.
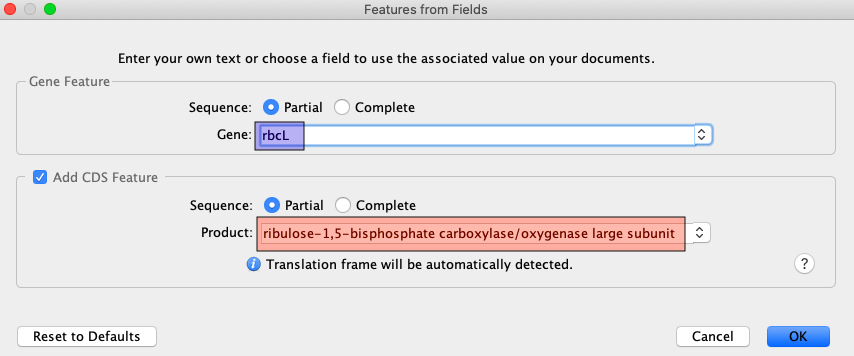
The following table shows the corresponding Gene and CDS Product name for various markers commonly used in DNA barcoding. Copy and paste directly from here, or look up existing sequences in GenBank to see the preferred notation for any protein-coding genes not listed here.
Gene |
CDS Product |
|---|---|
matK |
maturase K |
rbcL |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit |
CytB |
cytochrome b |
psbA |
Photosystem II protein D1 |
However, when creating submission files for sequences that are non-coding, such as the psba-trnH intergenic spacer or pseudogenes, follow the Annotating Sequences for GenBank Submission instructions.
Primers
When submitting from consensus sequences, the LIMS fields that hold the PCR primer names and PCR primer sequences should be populated automatically. Otherwise, it may be necessary to choose the correct fields from the dropdown menus in Geneious.

Confirming Submission
Once all fields have been appropriately populated in the Submit to GenBank window, click “OK” on the lower right of the window.
A Submission Warnings window will appear with three tabs.
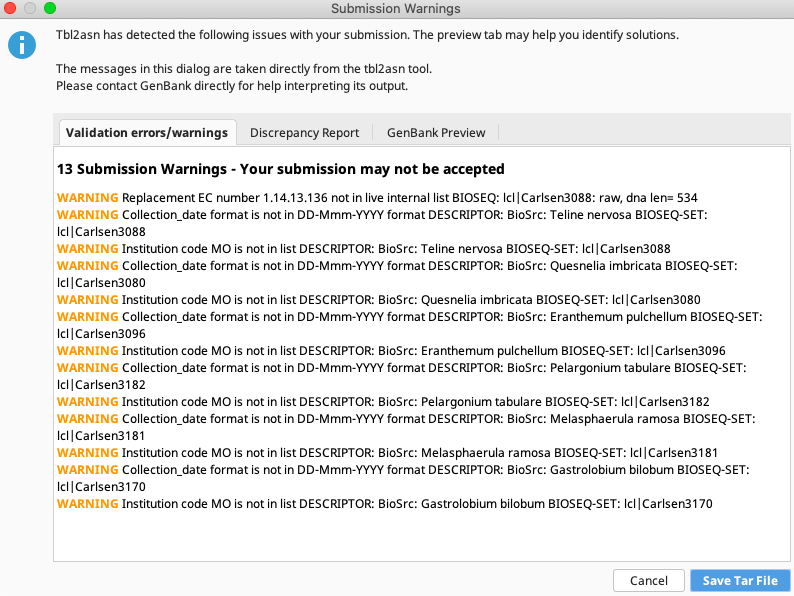
Address any warnings seen on the first tab “Validation errors/warnings” if they will result in the submission being rejected.
Warnings concerning date format or collection code (seen here) can generally be ignored. Also view the “GenBank Preview” tab to make sure all features and metadata will appear in GenBank records as expected.
If submitting through “Save a local file (.tar)”, once ready, click “Save Tar file” in the lower right corner and email the resulting zipped file to gb-sub@ncbi.nlm.nih.gov.
If submitting directly through “Upload New Submission”, once ready, click “Submit to GenBank” in the lower right corner and a BankIt submission which includes the sequences, annotations, and metadata will be sent to GenBank directly.
After Submission
A few days after submission, an email should be received from the NCBI admin team confirming the submission and assigning GenBank accession numbers to each of the sequences. Any issues that may have come up during post-submission processing will also be addressed.
Accession numbers should be reported back to the relevant departmental staff so they can be linked to the genetic sample records in EMu. If the sequencing project was funded by SIBN, please also report accession numbers back to the SIBN project manager.