Generating Extraction Plates
Creating the Plate (96 wells)
The first step of the LIMS workflow is the generation of a DNA extraction plate. In LIMS terms, an “extraction plate” is a set of DNA extractions following identical protocols and can include anywhere from a single extraction up to a plate of 384 extractions. This set of extraction will be moved through the entire pipeline together.
To create an extraction plate:
Click the “New Reaction” button on the Geneious Prime Toolbar.

In the dropdown menu of the New Reaction window, choose “Extraction” from the “Type of reaction” dropdown menu. Select the “96 well plate” size option and click “OK”.
A new window will open displaying a map of a 96 well extraction plate. The FIMS data must be uploaded to an empty extraction plate so that each specimen’s field data is correctly associated with that same specimen’s laboratory workflow.
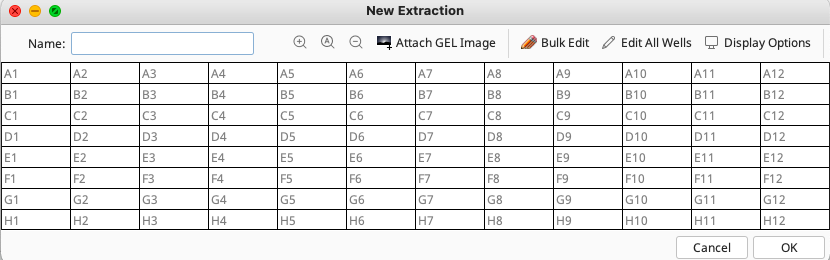
Insert a name for the extraction plate into the relevant field in the window. (See the LIMS Naming Conventions for best practices)
Click the “Bulk Edit” button.
A new Edit Plate window will appear. Within this window, LIMS can pull the GEOME FIMS metadata for all tissues to be eventually attached to each tissue’s LIMS workflow ID.
If creating a rerun extraction plate from existing LIMS extractions, continue with the Creating the Rerun Plate instructions.
If creating a new extraction plate with new Tissue IDs, continue below.
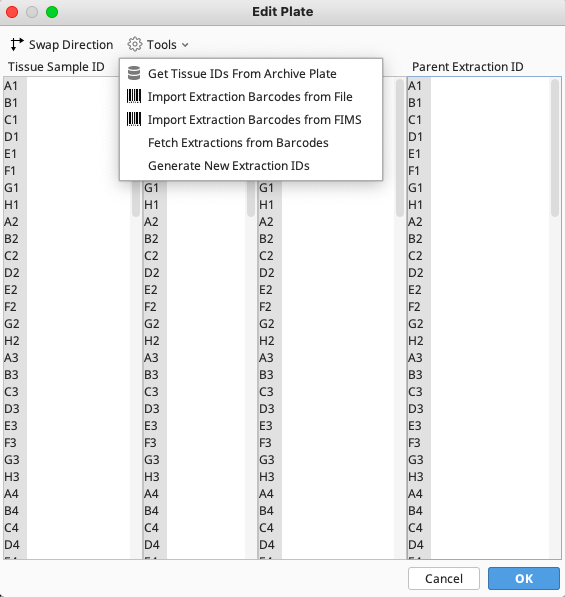
Click on “Tools” and select “Get Tissue ID’s From Archive Plate” from the dropdown menu.
Another new window Get FIMS plate will appear.
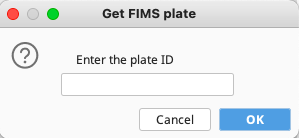
Here, enter the tissuePlate value that was included in the GEOME FIMS spreadsheet for this expedition. Click “OK” once complete.
A new window will appear indicating the LIMS is fetching the tissue ID’s from the FIMS database. When successful, the “Tissue Sample Id” fields in the Edit Plate window will be populated. Alternatively, if a message indicating that the plate cannot be found in the FIMS is recieved, check:
that the plate ID is spelled exactly as it was included in the GEOME FIMS spreadsheet and/or
that the FIMS spreadsheet was successfully uploaded to the GEOME FIMS.
After successful upload, select “Generate New Extraction IDs” from the “Tools” dropdown menu in the Edit Plate window. The LIMS will automatically populate the second column of the window (“Extraction ID”). The .1 indicates this is the first extraction in the LIMs for that particular tissue, .2 would indicate it is the second and so on.
Finally, import the Extraction Barcodes (the 2D barcode on the bottom of the matrix tubes) for each DNA extract. This can be done in two different ways:
Option 1: This is the recommended method. Select “Import Extraction Barcodes from FIMS” from the “Tools” dropdown menu. A new window will appear, and select the GEOME FIMS column that contains the extraction barcodes from the dropdown menu - typically this is “tissueOtherCatalogNumbers”. For this to work, ensure that the FIMS contains the unique extraction barcodes.
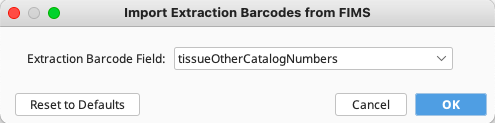
Option 2: Select “Import Extraction Barcodes from File” from the “Tools” dropdown menu. Select the file generated from the plate scanner. Ensure that the well order of the plate scan matches the well order displayed in the Edit Plate window.
Once this is complete Click “OK”. Clicking “OK” will attach the FIMS metadata to the new extraction plate.
Back on the New Extraction window, click on “Edit All Wells” to open a new window.
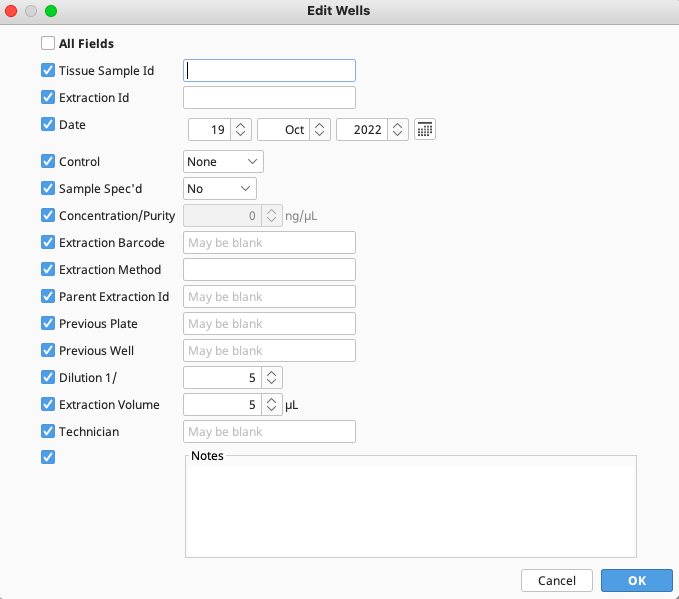
Enter information about the extraction - date, method, technician etc. into this window. Once complete, click “OK”.
Click “OK” in the New Extraction window for the LIMS to save the extraction plate.
Creating the Working Stock (96 wells)
After DNA extraction, the extracts are typically stored in archive plate for eventual storage in the biorepository while aliquots are kept in a separate plate to be used in PCR reactions. This protects the bulk of the DNA from freeze/thaw cycles as well as possible contamination during lab work. It is therefore considered best practice to create a working stock extraction plate in the LIMS to represent this aliquoted plate in the lab.
After the initial extraction plate has been created in the LIMS, it is easy to create a working stock from this same plate.
Search for the recently created extraction plate in the Biocode Plugin. See Searching for Existing Plates for instructions.
Highlight the plate and click “New Reaction” in the Genenious Prime Toolbar.
Make sure the box next to “Create plate from existing plate documents” is ticked, and that the “Type of reaction” is set to “Extraction”. Click “OK”.
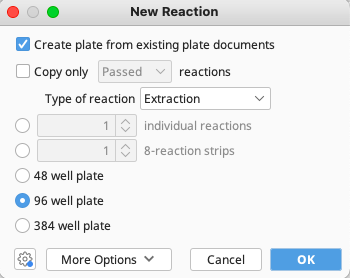
As before, a new window will open displaying a plate map, except this time all needed data should already be populated. Give the working stock a name, following the LIMS Naming Conventions.
Click on “Bulk Edit” to see the data that was pulled in from the extraction plate. The “Extraction Barcodes” column should be empty. Leave it empty. Re-entering Extraction Barcodes here will remove them from the parent plate.
The working stock plate extractions should be linked to the original extraction by the value that should be automatically populated in the “Parent Extraction ID” column. The values in the “Extraction ID” column should also automatically have increased.
Click “Save” to save the working stock plate to the LIMS.
Additional Information
The Edit Plate Window
In this window, the well locations are displayed on the left hand side of each column to make placement easier. The “Swap Direction” button allows the user to choose between reading the plate horizontally or vertically.
Under the “Tools” dropdown menu there are a number of options available.
- “Get Tissue IDs From Archive Plate”
This allows the extraction plate to be filled with extraction IDs from the FIMS data source.
- “Import Extraction Barcodes from File”
This allows the Extraction Barcode values to be directly imported from the output file of the scanner if 2D well barcodes are being used.
- “Import Extraction Barcodes from FIMS”
This allows the Extraction Barcode values to be directly imported from the FIMS.
- “Fetch Extractions from Barcodes”
This is used during “cherry picking” to populate newly reconstituted plates from prior plate locations if physically moving the extractions from original plate to the cherry picked plate. This can also be used to pull sample info if cherry picked plate is aliquoted, but remember to delete the extraction barcodes before saving cherry picked plate so the barcodes stay in their original locations in LIMS.
- “Generate New Extraction IDs”
This automatically generates appropriate Extraction IDs based on the Tissue Sample IDs.
Editing Wells
The “Edit All Wells” or “Edit Selected Wells” button, found in the top of the New Extraction window, opens an editor for LIMS data associated with each well. It is shown both when creating new plates, and when viewing existing plates in the database. Wells can be selected in the plate by dragging the mouse across the plate to select a number of wells, or holding down the shift and ctrl (command on mac) keys to help select multiple individual wells. When a well, or wells, have been selected, click “Edit Selected Wells” to edit data within those wells.
The Edit Wells window will open (see image above), and it has a column of checkboxes on its left hand side. Values in the checked fields will be applied to all selected reactions, and unchecked fields will be left as they are. Most values can simply be entered into a dialog box. Make sure to save the plate after making any edits to it.
Display Options
Clicking the “Display Options” button, found in the top of the New Extraction window, opens the Display window (below). The split-pane allows the user to choose any number of fields from the FIMS or LIMS database for display in the wells.
The available fields are shown in the left hand pane, and when fields are in the right hand pane they are displayed in the wells. To move a field between the two panes, select it and click the right or left arrow depending on the direction of the move. Once the fields to display have been decided upon, their display order can be altered using the up/down arrows on the right hand side of the dialog box. The field in the top position of the list will be displayed more prominently in the well, as it will be in larger, bold text.
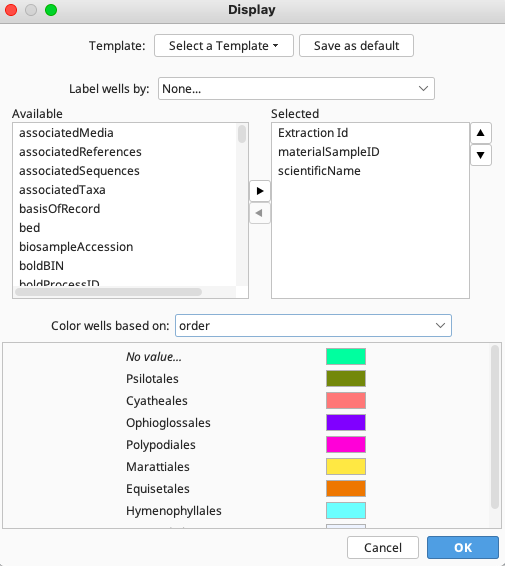
Each well can be color-coded according to a particular field value. To select the field for color-coding use the “Color wells based on” dropdown menu found at bottom of the dialog window. All possible values for that field will be displayed and a color can be assigned to each of the values using the color chart.
A display setting can be saved as a template by clicking the “Select a template” button at the top of the dialog window and clicking “Create template”. Click the “Save as Default” button to make that template the default. Separate defaults are stored for extraction, PCR, and cycle sequencing plates.