Exporting Sequence Metadata in TSV format
In Geneious Prime, gather all assembled contigs or consensus sequences from which metadata will be exported into a local folder.
Make sure the most up-to-date FIMS and LIMS metadata are associated with the sequences. To do this, follow the instructions on the Annotating with FIMS/LIMS Data page to annotate the sequence files with the current GEOME FIMS and Biocode LIMS data.
To export sequence metadata, first select all relevant files.
Export the sequence metadata by using the Export function in the Geneious Prime Toolbar and choosing “Export Documents”.
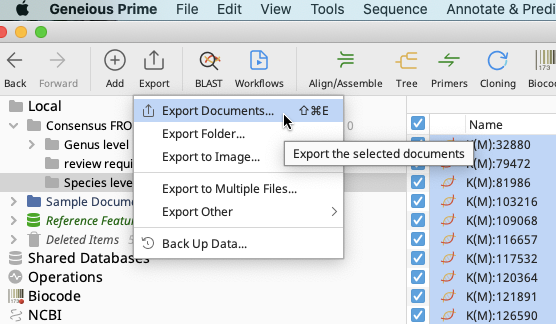
A Select Format window will open, but will contain different options dependent on whether export is from consensus sequence or contig files.
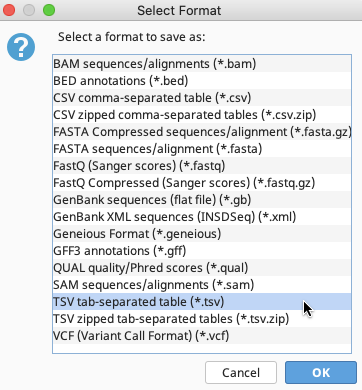
The Select Format window when exporting from consensus sequence files
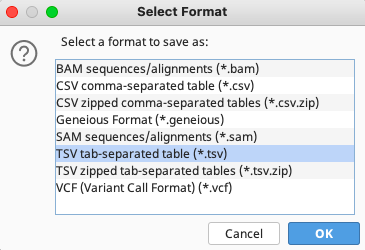
The Select Format window when exporting from contig files
In either case, select “TSV tab-separated table (*.tsv)”.
Next, name the fasta file, select the local directory in which to save it, and click “Proceed” on the Potential Data Loss window.

A list of FIMS and LIMS column headers should then appear. FIMS column headers start with a lowercase letter, and LIMS column headers start with an uppercase letter.
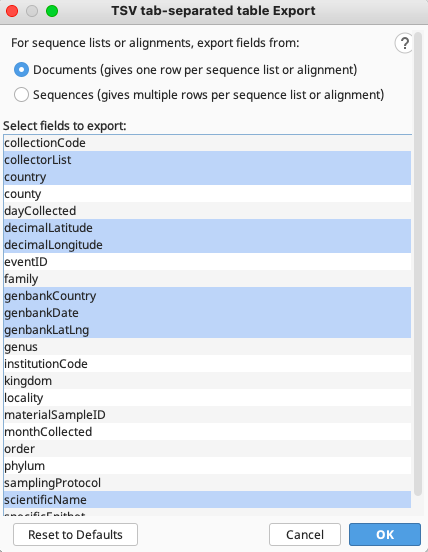
TSV tab-separated Export window that appears when exporting from contig files
If exporting from contig files, at the top of the window, ensure that “Documents” is selected. This will not be necessary if exporting from consensus sequence files.
From this list, choose all fields that are needed, then click “OK”. The tsv file will be exported to the location previously specified.