Populating the FIMS Spreadsheet
The GEOME FIMS Spreadsheet Template
The SI Barcode Network GEOME FIMS spreadsheet template is hosted on the SI Barcode Network GitHub page at https://github.com/SIBarcodeNetwork/SIBarcodeNetwork, called “SI Barcode Network FIMS template.xlsx”. (The direct link to download it is here.)
To personalize the columns included in the template, create a new account on GEOME and generate a customized template by following this link.
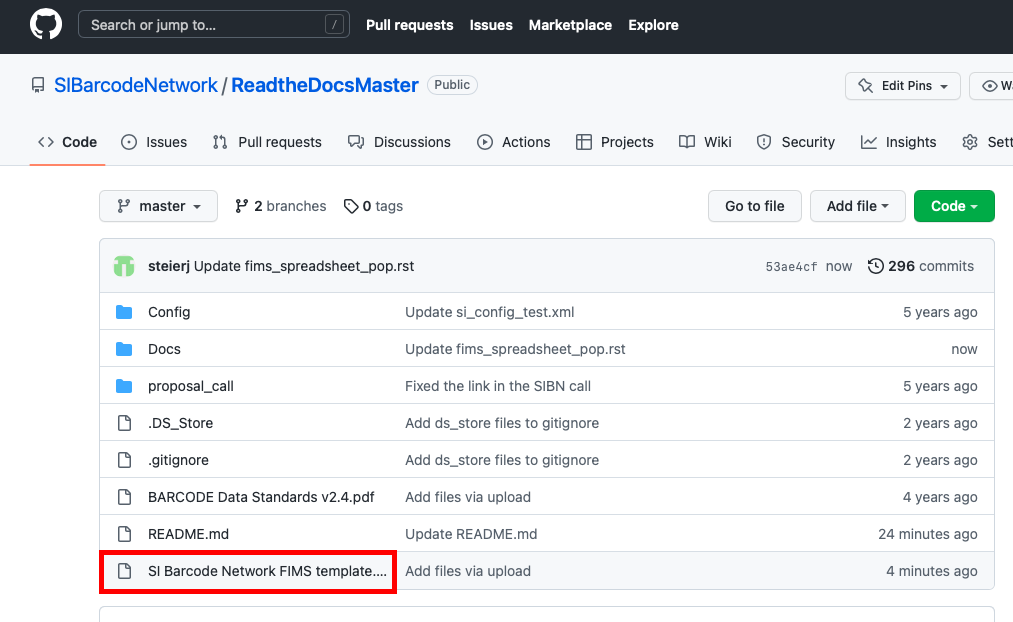
This screenshot of the SIBarcodeNetwork GitHub page shows the location of the specimen spreadsheet template.
It is a good idea to immediately rename this spreadsheet file with the dataset name according to the FIMS Naming Conventions.
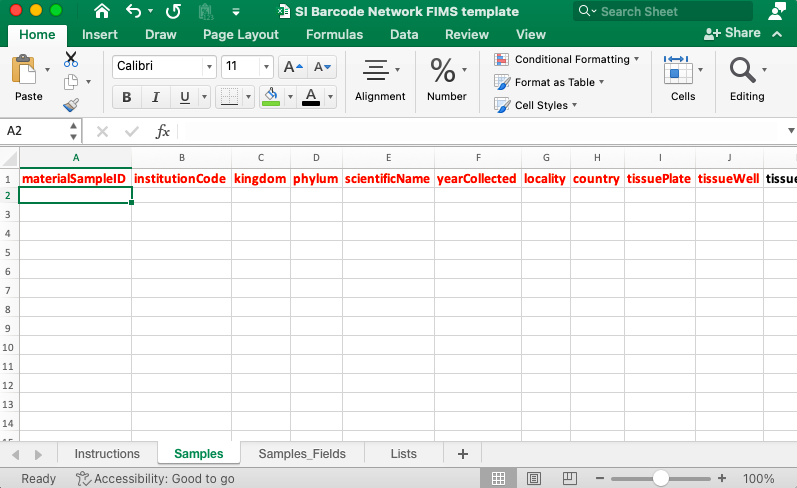
This screenshot shows a portion of the column headers of the specimen spreadsheet template.
There are four tabs in the spreadsheet: Instructions, Samples, Samples_Fields, and Lists. The Instructions tab explains how to use and populate the spreadsheet. The Samples tab is where data will be entered. The Samples_Fields tab includes definitions for all the columns on the Samples tab. The Lists tab includes all controlled vocabulary for certain fields.
Source of Columns
The GEOME FIMS allows projects to completely customize the fields that they use – along with the validation rules that accompany those fields. Since the goal of the SI Barcode Network is to get high quality sequences onto GenBank, the specimen fields in the SIBN FIMS template are limited to those that will end up in a GenBank record and those necessary for LIMS workflows. Fields are also based on DarwinCore fields, which should be mirrored in the field names. The SIBN subset of fields acts as a bridge between permanent collection databases (like NMNH EMu) and GenBank.
Column Colors
Red column headers indicate fields required before they can be uploaded to the FIMS.
Black column headers are not required, but they should still be filled out if the values are known.
Column Definitions
Column definitions are also found within the “Sample_Fields” tab of the template FIMS spreadsheet.
- materialSampleID
The collector’s specimen number. This number must be unique among the IDs within the expedition. Use the field number or voucherCatalogNumber, if no field number exists.
- institutionCode
This is the acronym for the institution or repository where the specimen voucher is stored. The GBIF Registry of Scientific Collections is a registry for all institution codes, and the institutionCode field will be validated against its list of codes. Vouchers accessioned at the Smithsonian will usually have the institutionCode “USNM”. Botanical vouchers stored in the US Herbarium will have the institutionCode “US”.
- collectionCode
The name, acronym, coden, or initialism identifying the collection or data set from which the record was derived. This collectionCode should be registered, along with the institutionCode, in The GBIF Registry of Scientific Collections.
- catalogNumber
A unique identifier (legacy) for the record within the data set or collection.
- voucherCatalogNumber
The catalog number used for the voucher specimen (assigned by host institution). Typically this field follows the format outlined in the specimen_voucher qualifier of the GenBank feature table and is structured as institution-code: (optional collection-code): specimen_id. Examples and a list of institutional abbreviations can be found on the INSDC page: https://www.insdc.org/controlled-vocabulary-specimenvoucher-qualifier.
- kingdom
The full scientific name of the kingdom in which the taxon is classified.
- phylum
The full scientific name of the phylum in which the taxon is classified. The list of phyla allowable in GEOME are taken from the Catalog of Life. ‘Unknown’ is an acceptable value. See controlled vocabulary in the “Lists” tab of the spreadsheet.
- class
The full scientific name of the class in which the taxon is classified.
- order
The full scientific name of the order in which the taxon is classified.
- family
The full scientific name of the family in which the taxon is classified.
- genus
The full scientific name of the genus in which the taxon is classified.
- specificEpithet
The full scientific name of the specificEpithet in which the taxon is classified.
- infraspecificEpithet
The full scientific name of the infraspecificEpithet in which the taxon is classified.
- scientificName
The full scientific name of the specimen. When forming part of an Identification, this should be the name in lowest level taxonomic rank that can be determined. Use “TBD” if currently unavailable and update later.
- identifiedBy
A list (concatenated and separated by the pipe (‘|’) symbol) of names of people, groups, or organizations who assigned the Taxon to the subject.
- collectorList
A list (concatenated and separated by the pipe (‘|’) symbol) of names of people, groups, or organizations responsible for recording the original Occurrence. The primary collector or observer, especially one who applies a personal identifier (recordNumber), should be listed first.
- yearCollected
The four-digit year in which the voucher was collected, according to the Common Era Calendar. (If unsure of the value and will never come across it, add ‘Unknown’, or if the data is currently unknown but will be known in the future, add ‘TBD’.)
- monthCollected
The two-digit numerical month in which the voucher was collected. This will be validated to being in the range from 1 to 12.
- dayCollected
The integer day of the month on which the voucher was collected. This will be validated to being in the range from 1 to 31.
- country
The name of the country or major administrative unit in which the Location occurs. This field will be validated against the INSDC country list (http://www.insdc.org/country.html). See controlled vocabulary in the Lists tab of the spreadsheet.
- locality
The specific description of the collection location. Less specific geographic information can be provided in other geographic FIMS fields (country, stateProvince, county, municipality, etc.). This term may contain information modified from the original to correct perceived errors or standardize the description. (If unsure of the value and will never come across it, add ‘Unknown’, or if the data is currently unknown but will be known in the future, add ‘TBD’.) This will be combined with the “country” FIMS field in the GenBank record.
- decimalLatitude
The geographic latitude (in decimal degrees, using the spatial reference system given in geodeticDatum) of the geographic center of a Location. Positive values are north of the Equator, negative values are south of it. Legal values lie between -90 and 90, inclusive.
- decimalLongitude
The geographic longitude (in decimal degrees, using the spatial reference system given in geodeticDatum) of the geographic center of a Location. Positive values are east of the Greenwich Meridian, negative values are west of it. Legal values lie between -180 and 180, inclusive.
- tissueType
A list (concatenated and separated) of the tissue types sampled from this individual, together with any tissue identifiers that were assigned to them
- tissuePlate
The name of the plate (typically a 96 well plate) containing the tissue subsamples that will be consumed for DNA extractions for projects.
- tissueWell
The well location in the tissue plate – formatted as follows: A01, A02, etc.
- tissueID
This is the unique identifier for the tissue sample from which the DNA was extracted. This identifier must be unique across all projects. Typically, this value is the materialSampleID + “.#” . This allows for multiple occurences within FIMS for a single specimen. If there are multiples of a tissue sample in different wells, please use the following format: materialSampleID.1, for the first occurence, materialSampleID.2 for the second occurence, and so on.
- tissueBarcode
Alphanumeric barcode given to tissue by storing institution. The NMNH Biorepository number may be stored here.
- tissueOtherCatalogNumbers
This is the 2D barcode of the storage tube which contains the DNA extract of the specimen. This field will not be populated until after the DNA extraction process is complete.
- tissueCatalogNumber
The catalog number used for the tissue specimen (assigned by host institution). Typically this field follows the format outlined in the bio_material qualifier of the GenBank feature table and is structured as institution-code: (optional collection-code): specimen_id. Examples and a list of institutional abbreviations can be found on the INSDC page: https://www.insdc.org/controlled-vocabulary-specimenvoucher-qualifier.
- boldProcessID
BOLD Process IDs are unique codes automatically generated for each new record added to a project within the Barcode of Life Database.
- boldBIN
Add boldBIN provided by BOLD for COI barcodes here. For example: BOLD:AAF0202.
- associatedSequences
A list (concatenated and separated) of identifiers (publication, global unique identifier, URI) of genetic sequence information associated with the specimen. GenBank accession numbers can be backfilled into this field.
- associatedTaxa
A list (concatenated and separated) of identifiers or names of taxa and their associations with the Occurrence. For example: “host”:”Quercus alba”.