Exporting Sequences in FASTA format
In Geneious Prime, gather all assembled contigs or consensus sequences to be exported into a local folder.
Make sure the most up-to-date metadata are associated with the sequences. To do this, follow the instructions on the Annotating with FIMS/LIMS Data page to annotate the sequence files with the current GEOME FIMS and Biocode LIMS data.
If contigs or consensus sequences need to be renamed, follow the Using the Batch Rename Function instructions.
To export the documents in FASTA format, first select all relevant documents, then follow one of the two options outlined below depending on file type.
To export from contig files: click on “Export” in the toolbar, then select “Export other” and “Consensus”.
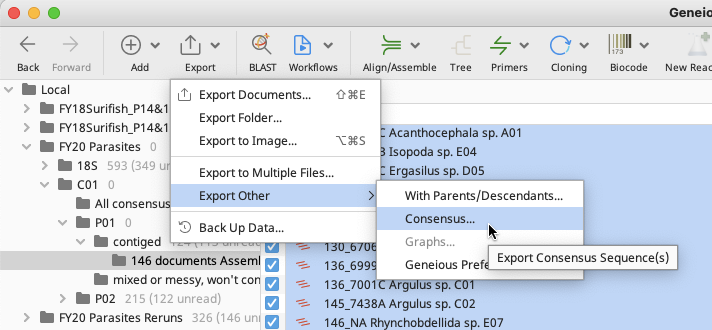
In the Consensus window that pops up, choose settings as appropriate or leave as defaults. Click “OK”.
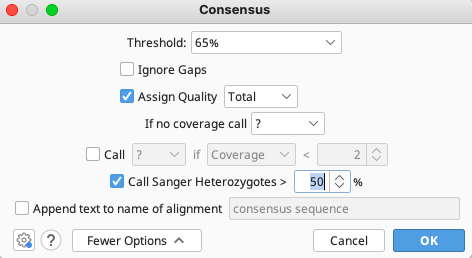
To export from consensus sequence files: click on “Export” in the toolbar, then select “Export Documents”.
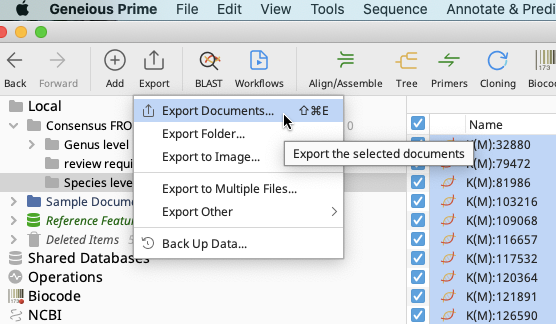
In the Select Format window that pops up, choose “Fasta sequences/alignment (*.fasta)”. Click “OK”.
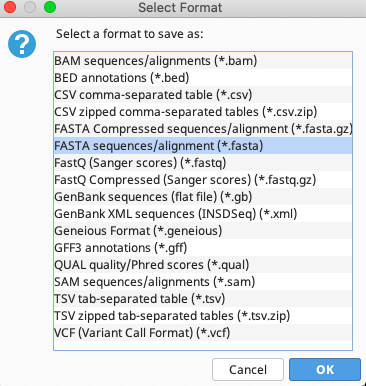
After naming the fasta file and selecting the local directory in which to save it, click “Proceed” on the Potential Data Loss window.

In the FASTA sequences/alignment Export window that appears, ensure that only the middle option “Replace spaces in sequence name with underscores” is checked. Uncheck the other two options. Choose Upper case or Lower case based on user preference.
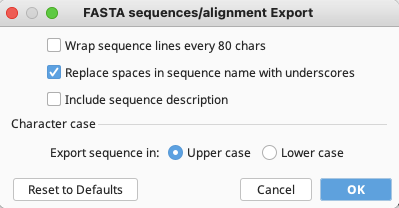
Click “OK” and the fasta file will be exported to the location previously specified.