Generating PCR Plates
Searching for Existing Plates
Biocode LIMS searches can return four types of documents:
Tissue sample documents – Each of these represents a tissue sample in the field database. Tissue documents contain collection and specimen information.
Plate documents – These represent a plate in the LIMS, and can be extraction, PCR, or cycle sequencing plate types.
Workflow documents – These contain a set of linked reactions performed on an extraction based on the marker sequenced.
Sequence documents - Sequences entered into the LIMS when assembled contigs are marked as pass/tentative/fail (see Reaction Status in LIMS).
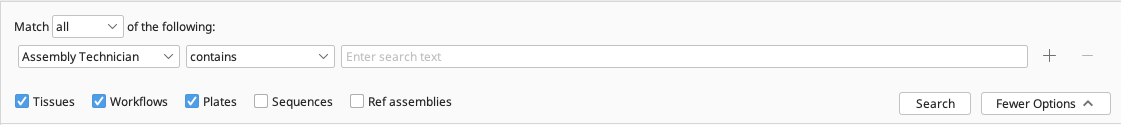
Searches are performed by entering text into the search box (found above the Document Table) and will return all matching documents.

Searches are restricted to particular types of documents by deselecting the checked boxes in the search window. To explicitly search against particular fields, click the “More Options” button. Click the “+” and “–” buttons on the right hand side to add or remove fields from the search. Choose the search fields of interest using the left-most dropdown menu and choose the middle dropdown menu to constrain search conditions.
Creating a PCR Plate from an Existing Extraction Plate
From this point forward, generating new plates, whether PCR or cycle sequencing, is very similar to the way the extraction plate is generated. Geneious can use an existing plate as a guide to create a new plate, so that new reactions are appended onto corresponding workflows.
To do this, select an existing plate from the Biocode LIMS plugin Search Results found in the Document Table.
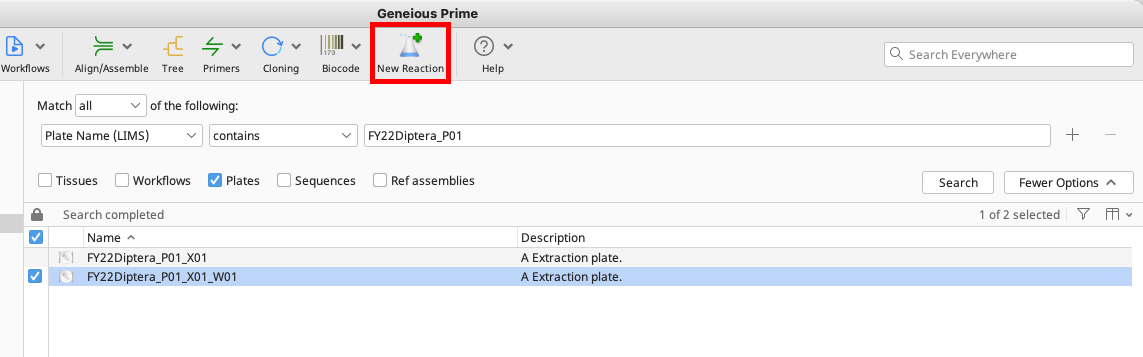
Click “New Reaction” in the Geneious Toolbar and check the “Create plate from existing document” checkbox.
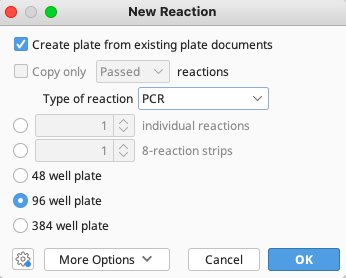
Select the “Type of reaction” as PCR, click OK and a New PCR window will appear.
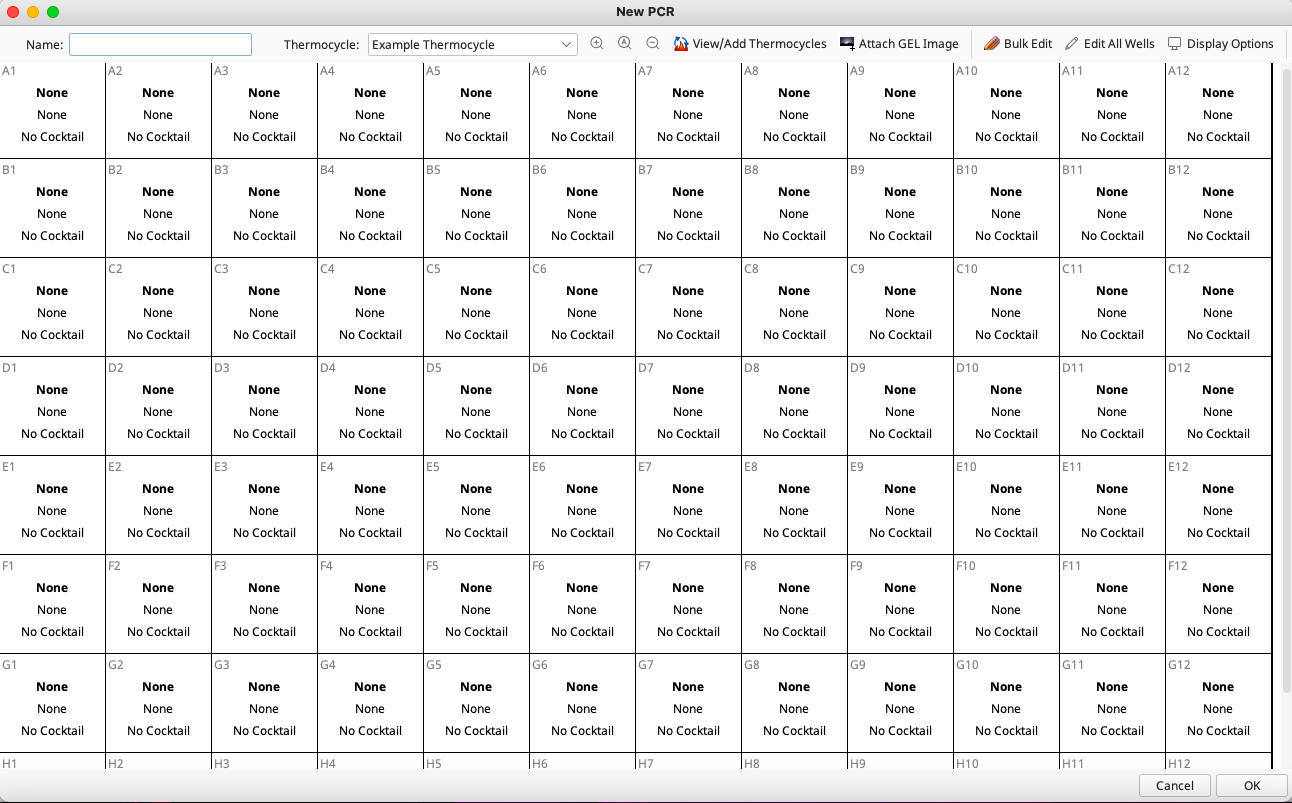
Enter a plate name in the box provided. This name should be descriptive and unique to allow easy and successful searching in the LIMS. (See LIMS Naming Conventions)
The next step is to select the thermocycling protocol that was used for this PCR plate. The Geneious LIMS stores all previously used thermocycling profiles. These are found in the “Example Thermocycling” dropdown menu on the top of the New PCR widow. Creating new profiles is described in the “Additional Information” section at the end of this document.
Next, gel images can be uploaded by clicking the “Gel Images” button in the New PCR window and pointing to the relevant file on the local disk. More information on attaching a gel image and scoring is available in the “Additional Information” section below.
Lastly, to edit all wells on the PCR plate, select “Edit All Wells”. Alternatively, to edit individual wells, highlight the selected wells in the plate and click “Edit Selected Wells” on the top of the New PCR window. In both cases the Edit Wells window opens.
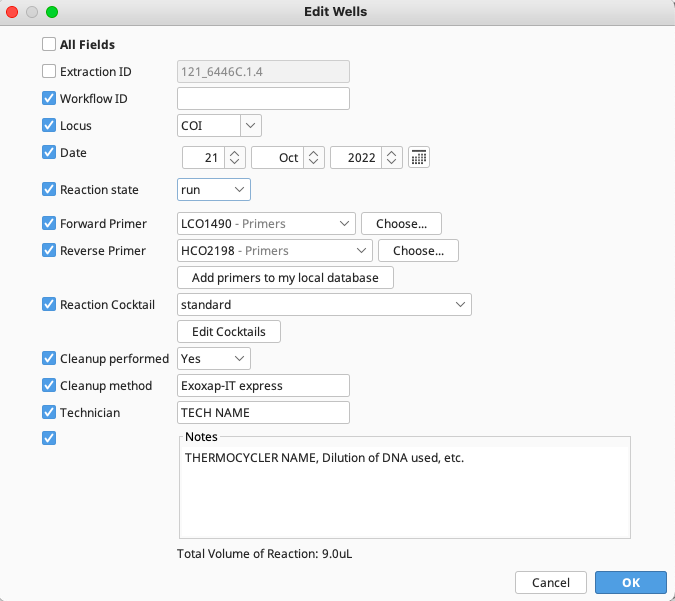
This window has a column of checkboxes on its left hand side. Values in the checked fields will be applied to all selected reactions, and unchecked fields will be left as they are.
The target locus can be selected in the “Locus” dropdown list. This list is pre-populated, however, it can be manually edited to include additional loci for use in the LIMS. This value is essential for creating the Workflow ID for that PCR reaction.
PCR reactions can be marked as “not run” or “run” in “Reaction state” dropdown list. If making a judgement call on the amplification success of the PCR reaction based on a gel, the wells can be further marked as “passed”, “suspect”, or “failed”.
The forward and reverse PCR primers are selected using the “Choose” button which points to a selected folder on the Local Directory where primers are stored by the user. (See Creating Primers)
The “Reaction Cocktail” can be chosen from a list of existing cocktails, or by creating a new cocktail, described in the “Additional Information” section at the end of this document.
Indicate if PCR clean up was performed and what method - this will typically be “Exosap-IT” or “Exosap-IT express”.
Add any additional comments in the “Notes” section, including which thermocycler was used, what concentration the template DNA was, etc.
Click “OK” and the Edit Wells window will close, applying entered data to all selected wells.
Once all editing of the plate is complete, Click “OK”.
Additional Information
Creating Thermocycling Protocols:
To create custom thermocycler profiles, click “View/Add Thermocycles” in the New PCR window.
The Edit Thermocycles window opens (below) and this displays all of the stored profiles in the left section of the window and a diagram of the selected profile in the right section.
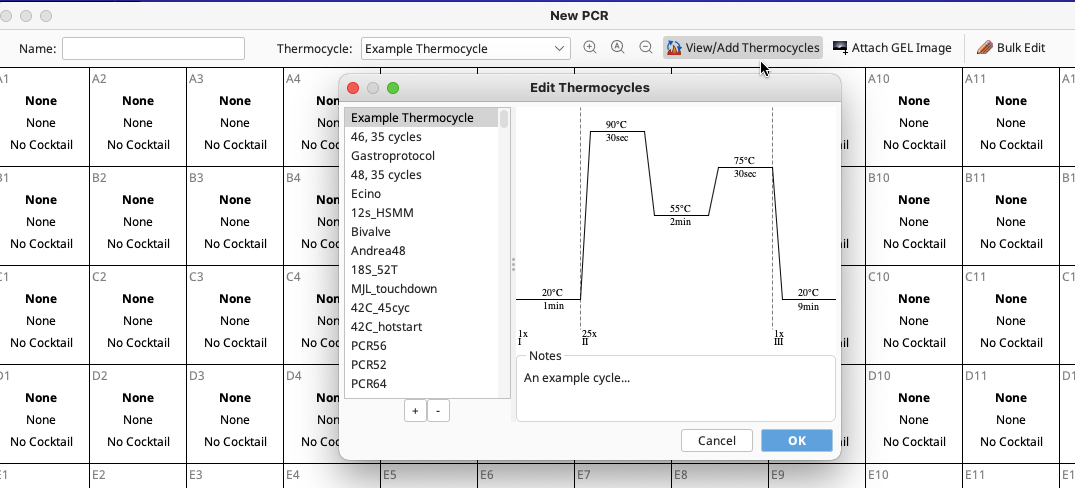
Click the “+” button on the lower left-hand corner of the window to add a new profile.
The New Thermocycle window will open. The temperatures and cycles associated with the new program can be entered using the dialog boxes and “Edit Cycles” buttons.
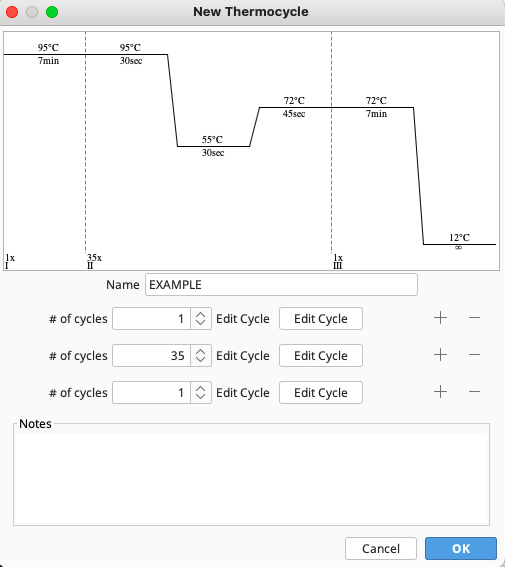
The “+” and “-” buttons to the right of the “Edit Cycle” button allow for addition or removal of steps in the thermocycling profile.
Note
It is not possible to modify thermocycler profiles once they are created and profiles can only be deleted if they are not associated with a workflow. Thermocycler profiles are also available to all users. As a result, double check that the required profile doesn’t already exist prior to creating it.
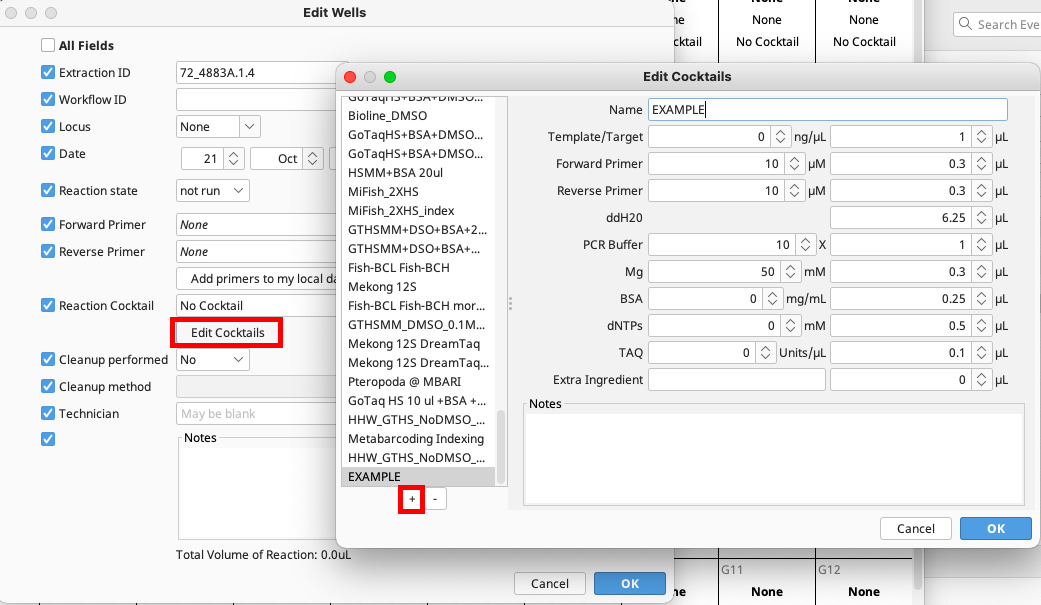
Creating Custom PCR Cocktails:
In the Edit Wells window, click “Edit Cocktails”, then click the + button in the dialog, and enter the volumes and concentrations in the fields provided.
There is space to store one extra ingredient (both concentration and volume) that isn’t in the provided ingredient list.
Any additional information about the new cocktail can be stored in the “Notes” field.
Note
It is not possible to modify cocktails once they are created, and cocktails can only be deleted if they are not associated with a workflow. Cocktails are available also to all users. As a result, double check that the required cocktail doesn’t already exist prior to creating it.
Attaching and Scoring Gel Images:
If attaching a gel (or gels) when creating a new plate click the “Attach GEL Image” button found on the top of the New PCR window.
If attaching a gel or gels to an existing PCR plate, when viewing the plate, click “GEL Images” at the top of the “Plate” tab in the Document Viewer.
In both cases the Edit Gel Images window will appear. Click “Add”, then browse to find the image or image files to be attached. Geneious Prime will accept images in JPEG, GIF, PNG, and TIFF formats. Notes can also be added each gel image at this point.
Once the image or images are attached, click the “Split GEL” button located above the image in the Edit Gel Images window.
To map the samples on the gel to the workflows in the plate:
Populate the four dialog boxes to the right and choose the read direction of the plate i.e., vertical or horizontal.
To generate the grid, drag the mouse/pointer over the image. If the grid is misplaced, start again by dragging the mouse over the image.
If the gel image is not straight causing the grid to line up incorrectly, use the sliding bar in the top right of the window to level the image.
Automated calling of pass/fail reactions in the gel is possible by checking “Automatically Score plate”. Click “OK” once the grid is in the correct position.
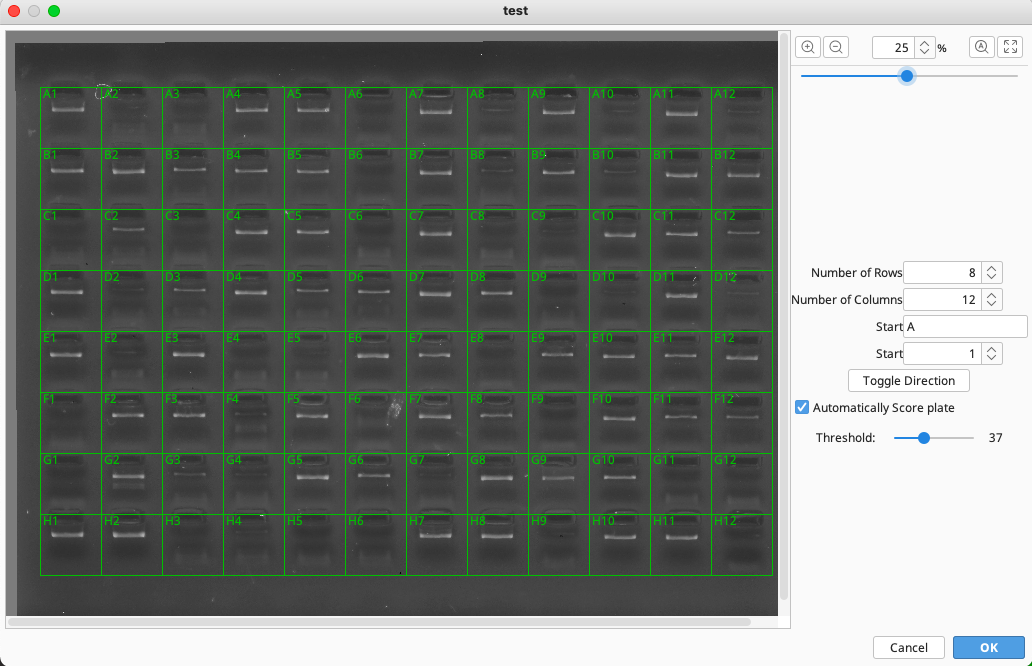
Back on the plate view, wells should be color coded according to the automatic scoring done on the gel image (green = pass, red = fail).
It is possible to manually change the call by double-clicking the well in the plate viewer, opening the Edit Wells window and changing the “Reaction state” from the dropdown menu.
The program may score primer dimers in the gel, so if they are present, extra care should be taken and the calls double checked.
